PLoS Genetics 8 (4): e1002641. doi:10.1371/journal.pgen.1002641
Patterns of Ancestry, Signatures of Natural Selection, and Genetic Association with Stature in Western African Pygmies
Jarvis, Joseph P., Laura B. Scheinfeldt, Sameer Soi, Charla Lambert, Larsson Omberg, Bart Ferwerda, Alain Froment, Jean-Marie Bodo, William Beggs, Gabriel Hoffman, Jason Mezey, Sarah A. Tishkoff.
Abstract
African Pygmy groups show a distinctive pattern of phenotypic variation, including short stature, which is thought to reflect past adaptation to a tropical environment. Here, we analyze Illumina 1M SNP array data in three Western Pygmy populations from Cameroon and three neighboring Bantu-speaking agricultural populations with whom they have admixed. We infer genome-wide ancestry, scan for signals of positive selection, and perform targeted genetic association with measured height variation. We identify multiple regions throughout the genome that may have played a role in adaptive evolution, many of which contain loci with roles in growth hormone, insulin, and insulin-like growth factor signaling pathways, as well as immunity and neuroendocrine signaling involved in reproduction and metabolism. The most striking results are found on chromosome 3, which harbors a cluster of selection and association signals between approximately 45 and 60 Mb. This region also includes the positional candidate genes DOCK3, which is known to be associated with height variation in Europeans, and CISH, a negative regulator of cytokine signaling known to inhibit growth hormone-stimulated STAT5 signaling. Finally, pathway analysis for genes near the strongest signals of association with height indicates enrichment for loci involved in insulin and insulin-like growth factor signaling.
Link (Free Full Text PDF)
Human Biology 84 (1):11-43. 2012 doi: http://dx.doi.org/10.3378/027.084.01
Changing Language, Remaining Pygmy
Bahuchet, Serge.
Abstract
In this article I am illustrating the linguistic diversity of African Pygmy populations in order to better address their anthropological diversity and history. I am also introducing a new method, based on the analysis of specialized vocabulary, to reconstruct the substratum of some languages they speak. I show that Pygmy identity is not based on their languages, which have often been borrowed from neighboring non-Pygmy farmer communities with whom each Pygmy group is linked. Understanding the nature of this partnership, quite variable in history, is essential to addressing Pygmy languages, identity, and history. Finally, I show that only a multidisciplinary approach is likely to push forward the understanding of African Pygmy societies as genetic, archeological, anthropological, and ethnological evidence suggest.
The mystery of the Pygmy phenotype continues to stimulate scientific research. The two recent papers tackle it differently: Jarvis et al. (2012) report on possible genetic loci that are target of natural selection in Pygmies; Bahuchet (2012) summarizes gives an interdisciplinary overview of the Pygmy problem and recapitulates the evidence for an ancient forager substratum in Pygmy languages. It’s generally believed that Pygmies, who practice hunting and gathering, descend from ancient tropical foragers that came in contact with Bantu and other agricultural populations in the last few thousand years as agriculturalists expanded across central and southern Africa. Pygmies speak languages closely related to Bantu, Ubangian (both part of Niger-Congo) and Central Sudanic (Nilo-Saharan) languages. In the map below the Bantu-speaking Pygmies are green, the Ubangian-speaking ones are purple and the Central Sudanic speaking ones are red. The population numbers are from Hewlett B. S., and J. M. Fancher. “Central African Hunter-Gatherer Research Traditions,” in Oxford Handbook of the Archaeology and Anthropology of Hunter-Gatherers. Oxford, 2011.
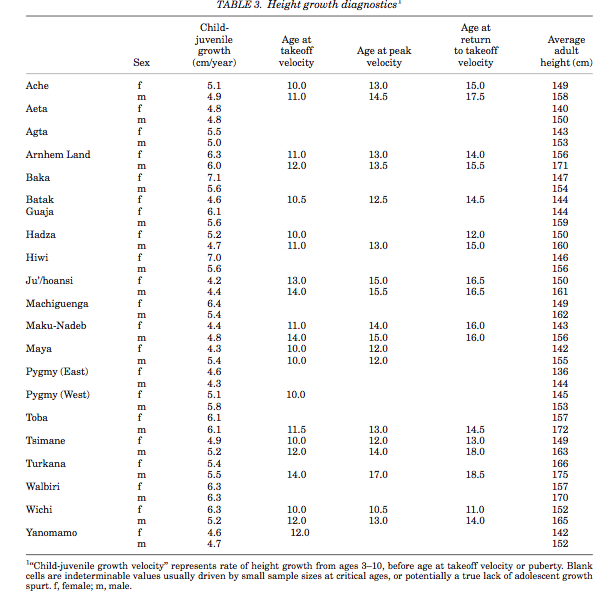
There are 12 Pygmy groups in total. None of them speak a language of their own. It’s assumed that all Pygmies lost their original languages and adopted the languages of their agricultural neighbors. Languages of all the widely dispersed and tropical forest-bound foraging groups disappeared; meanwhile, the languages of other African foragers such as Hadza, Sandawe and San not only remained intact but in fact influenced the languages of incoming agriculturalists by sharing their unique clicking sounds with some of them. Pygmy kinship systems and terminologies are also similar to those of Bantu, Ubangian and Central Sudanic peoples: they belong to various forms of derived Lineal and Generational types and don’t demonstrate anything reminiscent of the truly archaic symmetrical kinship systems that Alan Barnard and others described among Khoisan-speaking hunters and gatherers that have parallels in Australia and the New World. The issue, therefore, is not with Pygmies’ foraging lifestyle – foragers globally are capable of not only preserving their original languages but impacting the languages of agriculturalists. The issue is with the Pygmy phenotype: they are ~17 cm shorter on average than their agricultural neighbors and among the shortest populations globally (see Table 3 below, from Walker et al. “Growth Rates and Life Histories in Twenty-Two Small-Scale Societies,” Am J Hum Bio 18 (2006), p. 301).
Pygmies also have a very short life-span (between 15.6 and 24.3 years). Other explicit biodemographic metrics seem to be no different from other human populations. The origin of short stature in African Pygmies (and among other pygmoid populations globally) remains a subject of debate and research. The antiquity of Pygmy phenotype remains without a direct proof. As Bahuchet (p. 32) notes,
“Anthropological remains are very rare in Central Africa, and only three sites delivered some bones. The oldest concerns 18 individuals excavated in the Shum Laka shelter (NorthWest Cameroon) in three tombs from 6,000 years BP and six more dated circa 15th century CE. Along the Ubangi River, a single skull was found dated circa 1st century BC. The most recent is a skeleton from the Matangai Turu rock shelter in Ituri (DRC), dated 1235 AD. None of these remains can be clearly attributed to some morphotype, either Pygmy or not.”
Geneticists have argued, on the basis of mtDNA, nuclear and other genetic systems, that Pygmies have some of the most divergent human genetic lineages. They date these lineages at 70-100,000 BP, which places the origin of Pygmies during the African Middle Stone Age, or 30,000 plus years prior to the onset of Upper Paleolithic in Eurasia. However, there’s no evidence for Pygmy phenotype among Middle Stone Age African anatomically modern humans or archaics.
Unlike San who tend to claim a distinct place of their own in mtDNA phylogenies and differ from the rest of extant humans, both African and non-African, Pygmies are not genetically isolated. This can be seen on the following tree of nuclear haplotypes (from Tishkoff et al. 2009):
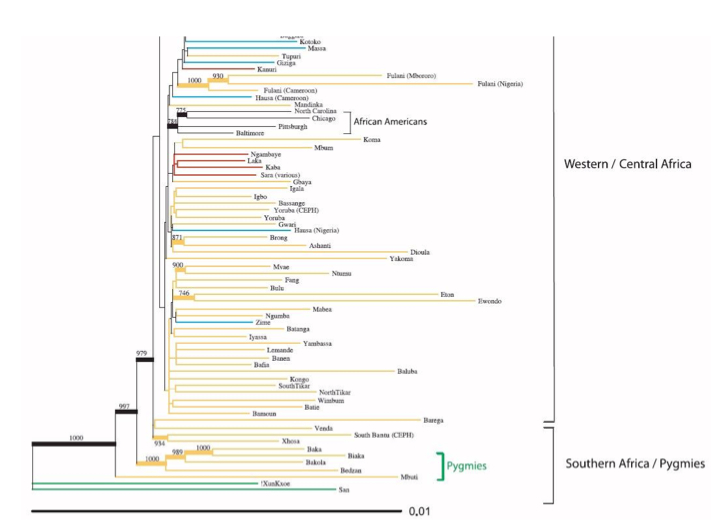 In fact, they share mtDNA lineages with their Bantu neighbors and not only because of admixture, but because of common descent. The Table below on the left clearly shows that Bakola, Biaka and Mbezele Aka have mtDNA hg L1c, while its sister hg L1b is found among agriculturalists (who also have a variety of other L lineages not attested in those Pygmy populations). The frequencies are indeed dramatically different, which is consistent with a bottleneck followed by isolation for Pygmies, but there is no question that Pygmies and Bantu are genetically sister groups. The phylogeny on the right (from Quintana-Murci et al. 2008) shows that Western Pygmy (in yellow) and Bantu mtDNA lineages (in blue) are thoroughly interspersed, which is consistent with the linguistic evidence of recent common ancestry between them.
In fact, they share mtDNA lineages with their Bantu neighbors and not only because of admixture, but because of common descent. The Table below on the left clearly shows that Bakola, Biaka and Mbezele Aka have mtDNA hg L1c, while its sister hg L1b is found among agriculturalists (who also have a variety of other L lineages not attested in those Pygmy populations). The frequencies are indeed dramatically different, which is consistent with a bottleneck followed by isolation for Pygmies, but there is no question that Pygmies and Bantu are genetically sister groups. The phylogeny on the right (from Quintana-Murci et al. 2008) shows that Western Pygmy (in yellow) and Bantu mtDNA lineages (in blue) are thoroughly interspersed, which is consistent with the linguistic evidence of recent common ancestry between them.
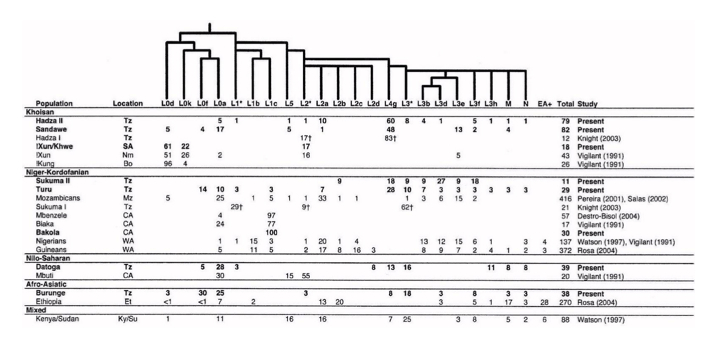
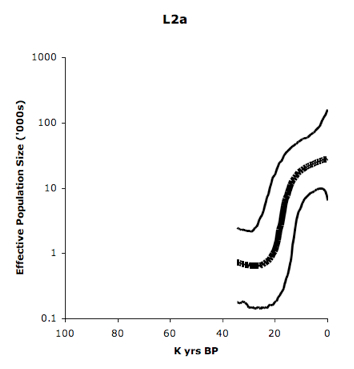
Eastern Pygmies (Mbuti) who speak the Bantu Bila language have 65% of hg L2a2. Again, L2 lineages are abundantly found among Niger-Congo speakers, as the Table above on the left demonstrates. L2a is considered to contain one of the strongest mtDNA signals of Bantu expansion as reflected in its steep expansion curve (see below, from Atkinson et al. 2009, Suppl Mat, Fig. S2).
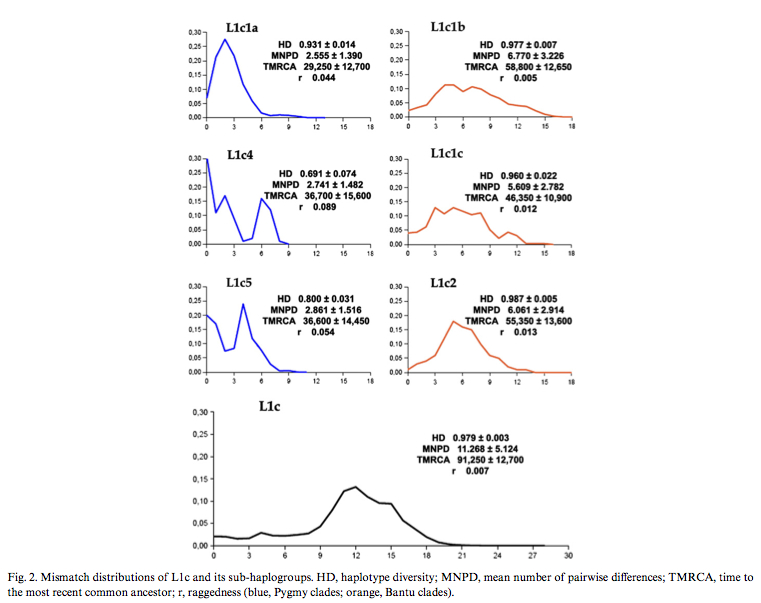
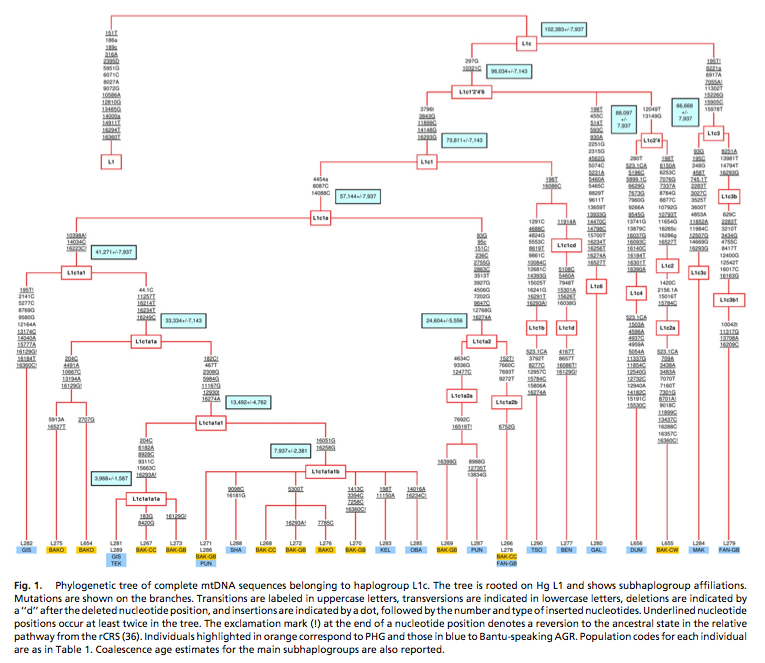
Batini et al. (2007) published (see Table 2 below) diversity values and coalescent dates for Pygmy and Bantu subclades of mtDNA hg L1c. Bantu subclades prove to be older and more diverse than the corresponding Pygmy subclades. Mismatch distributions are different between the forager subclades and the farmer subclades, which is consistent with different demographic histories, but nothing indicates that Pygmies are “older” than Bantu.
Although the myth of twelve lost languages of the Pygmies is strong and wide-spread, linguistic classifications are in no way discrepant with genetic evidence: Pygmies are closely related to Bantu both linguistically and genetically. One may argue about the timing of splits among Niger-Congo languages, but it seems rather unlikely that these splits occurred during the Middle Stone Age. Mainstream historical linguistics dates the break-up of the Niger-Congo phylum in the Holocene in line with the near-universal contention that comparative method doesn’t extend much beyond 5,000 years BP. To make the linguistic dates for the separation of the Niger-Congo languages comparable to the molecular dates for the Pygmy-Bantu split will require the adoption of a version of Paleolithic Continuity theory, which is rejected by mainstream linguistics. Unless Pygmies and Bantu are related twice, first genetically and then linguistically, which is nonsensical, one is left with the conclusion that Pygmy populations are not as ancient as geneticists portray them. Again, it’s the peculiarity of the Pygmy phenotype that conjures up an image of primordial people in scholars’ minds. Had Pygmies been of normal stature, they would looked like an offshoot of Niger-Congo and Central Sudanic populations representing either a slightly earlier, pre-agricultural wave of expansion into the tropical forest, or, as Roger Blench (“Are the African Pygmies an Ethnographic Fiction?” in Challenging Elusiveness: Central African Hunter-Gatherers in a Multidisciplinary Perspective. Pp;. 41-60. Leiden, 1999) suggested, a specialized caste of hunters-gatherers similar to the Numu blacksmiths castes of West Africa. Bahuchet (see Table 5 below on the left from Bahuchet 2012 and Table 1 below on the right, from Bahuchet S. “Historical Perspectives on the Aka and Baka Pygmies in the Western Congo Basin,” in Proceedings of the 86th Annual Meeting of the American Anthropological Association, November 19, 1987. Chicago, p. 8) has collected over the years a massive corpus of specialized vocabulary items shared across two Pygmy groups (Baka and Aka) that purportedly represents retentions from a foraging substratum in the Pygmy languages.
One can observe different patterns in this data: there are lexical items that are shared between foragers and agriculturalists that belong to the same language family (e.g., zumbi/zombi in Aka and Ngando, kopa ‘ax’ in Baka and Ngbaka); some items are different between a forager language and its agriculturalist counterpart but they are not shared with the other forager language either (e.g., mbenga ‘spear’ in Baka); others are shared between two agriculturalist languages and one forager language (e.g., nzoi ‘honey bee’ in Aka, Ngando and Ngbaka). Finally, a number of items are indeed shared by Aka and Baka to the exclusion of Ngando and Ngbaka (e.g., nguia vs. pame ‘Potamochoerus’). The shared Aka/Baka vocabulary is mostly concentrated in the “natural history” and “common technics and tools” classes, which is consistent both with the foraging substratum language model (Bahuchet calls this language Baakaa) and the Pygmies-as-caste model.
Jarvis et al. (2012) provide evidence that admixture between Bantu and Western Pygmies has created a genetic trace that’s radically different from the one seen in a more recent admixed population, African Americans:
“…The very short average tract lengths of inferred ancestry we observe (average Bantu tract length of 3.1+/24.6 Mb) are strikingly different from those seen in simpler admixture scenarios (e.g. African Americans). The shorter average Bantu tract length we observe appears to reflect the long history of admixture between Western Pygmy and neighboring Bantu populations that has taken place, possibly since the Bantu expansion into the rain forest several thousand years ago.”
On the surface, this appears to confirm the possibility of a massive language shift in Pygmy communities. Indeed, the towering Bantu intruders had plenty of time to impose their languages on miniature foragers. But naive sociolinguistics aside, the truth of the matter is that language shifts don’t require thousands of years to complete. A language shift may be complete in a few hundred years or it may be on the way but then cease. What Jarvis et al. found, instead, is evidence for prolonged co-existence and mutual dependence between Bantu agriculturalists and Pygmy foragers, precisely the kind one would expect if Pygmies were a caste. Long-term co-dependency of foragers and agriculturalists also points to the possibility of social or sexual selection to operate between the two groups, with Bantu agriculturalists favoring taller Pygmy women as wives for several millennia. Bahuchet (2012, p. 31) reports a number of terms of affinity and marital alliance shared between Pygmies and Bantu (brother-in-law: beˆndeˆ; jealousy for love: -koˆmbe`; son-in-law and courtship: –koˆpe`; pay for bridewealth: se`-; kinship through women: mobila).
Jarvis et al. (2012) seem to have found the genetic loci that are subject to selection in African Pygmies. Their study does not indicate that there was just one single selective pressure that affected a single locus in a proto-Pygmy, Middle Stone Age population and drove it purposefully toward short stature. Instead, it raises the possibility that Pygmies were subject to a number of selection events spanning multiple loci and that short stature is just a surface by-product of selection for traits other than stature, including “early reproduction, metabolism, and immunity, and that the functional variants (possibly regulatory in nature) that are targets of selection may have pleiotropic effects” (p. 8). This further points to a possibility that short stature is a product of convergent evolution in different parts Africa (and elsewhere in the world), in full accordance with linguistic classifications assigning each Pygmy language to a specific subfamily among Bantu, Ubangian and Central Sudanic languages. What is most remarkable is that Jarvis et al. (2012) identified a potential selection event that affected the genomes of all Africans, Pygmies included, and Africans only.
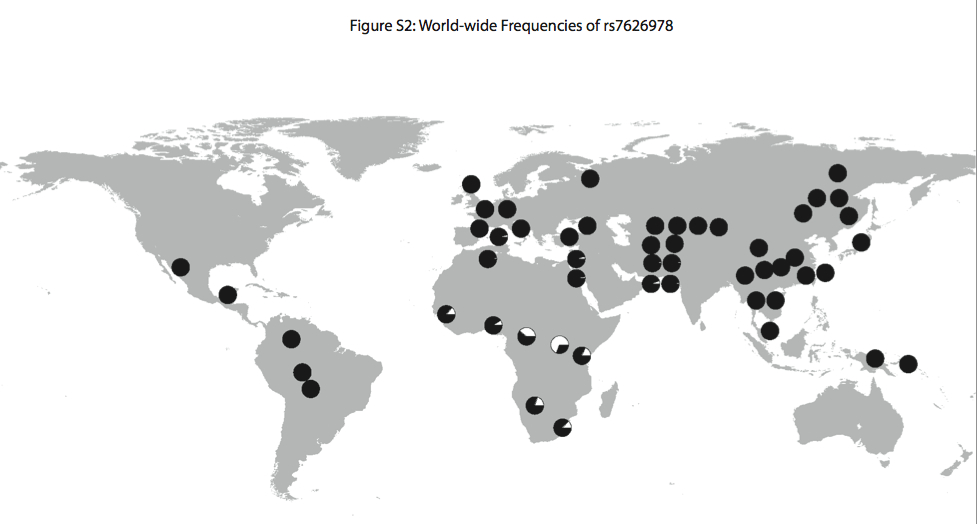
“Interestingly, the highest FST SNP in the region for which there is genotype data from the CEPH human diversity panel (rs7626978 at position 48505831; FST = 0.53) shows a striking global distribution in which the minor allele is most common in the Western and Eastern Pygmy, San, and neighboring populations and is nearly absent outside of Africa (Figure S2).”
Could it be that selection accounts for African genetic divergence and manifests itself across multiple genetic systems, including mtDNA?